Contents
% bpdq_demo_1d : Demo for BPDQ reconstruction in 1-d % % This demo first creates a random, sparse 1-d signal, creates a gaussian % random sensing matrix, and computes the quantized measurements. % % Then the BPDQ algorithm is run to reconstruct the signal from the quantized % measurements. % % For Comparison, BPDN (corresponding to p=2) is also run, then the results % from both reconstruction programs are compared. % % Plots of relative errors ( ||x_n - x_(n-1)||/||x_n|| ) % as a function of iteration number are given to demonstrate empirical % convergence of the douglas-rachford algorithm used to compute the % BPDQ program. % % This demonstrates the method described in % "Dequantizing Compressed Sensing : When Oversampling and Non-Gaussian % Constraints Combine" % by Laurent Jacques, David Hammond, M. Jalal Fadili % ( submitted to IEEE Transactions on Signal Processing) % % This file is part of BPDQ Toolbox (Basis Pursuit DeQuantizer) % Copyright (C) 2009, the BPDQ Team (see the file AUTHORS distributed with % this library) (See the notice at the end of the file.) % Note: With parameters as set in this demo, execution time was 18 seconds % on 2.4 GHz Intel Core 2 Duo processor.
Experiment Setup and Demo Parameters :
- N - signal dimension
- K - signal sparsity
- M - number of measurements
- p - BPDQ moment (p=2 corresponds to standard BPDN)
- nbins - desired number of quantization bins
Note: Quantization threshold alpha is computed depending on nbins and on the signal instance)
N=1024; K=16; M=K*25; p=4; nbins=10;
Parameters for controlling BPDQ algorithm.
The Douglas-Rachford algorithm parameters * dr_lambda must be between 0 and 2, dr_gamma must be positive. * dr_gamma and dr_lambda are described in Jacques et. al. section IV-A; dr_lambda corresponds to alpha_t in equation 10
Note : a priori these parameters could vary at each iteration, in this code they are kept constant.
dr_verbosity and fca_verbosity control output of information about speed of convergence at each iteration, set to 0 to supress output, to 1 to show output. See: bpdq_d_r_iter.m and bpdq_prox_f_comp_A.m for details.
Other options * dr_maxiter, dr_relerr_thresh, fca_maxiter, fca_relerr_thresh are defined in in control_params in bpdq_1d.m, see bpdq_1d.m for details.
bpdq_opts={'dr_gamma',.05,'dr_lambda',1.5,'dr_verbosity',0,'fca_verbosity',0};
Create signal and sensing matrix
fprintf('Generating signal of length N=%g and sparsity K=%g\n',N,K); x=bpdq_generate_1d_signal(N,K); fprintf('Generating sensing matrix for M=%g random measurements\n',M); A=bpdq_generate_sensing_matrix(M,N);
Generating signal of length N=1024 and sparsity K=16 Generating sensing matrix for M=400 random measurements
Compute measurements and quantize
fprintf('Computing quantized measurements with %g quantization levels\n',nbins);
y=A*x;
alpha = 2.01*norm(y, Inf)/nbins;
yq = bpdq_quantize(y, alpha);
Computing quantized measurements with 10 quantization levels
Call BPDQ decoder
% Let's first compute apropriate epsilon (ref Jacques et. al section III-C) epsilon=bpdq_err_p(p,alpha,M); fprintf('Running BPDQ reconstruction with p=%g\n',p); [x_bpdq,D_bpdq] = bpdq_1d(yq,A,epsilon,p,bpdq_opts{:});
Running BPDQ reconstruction with p=4
Compare to BPDN
epsilon2=bpdq_err_p(2,alpha,M);
fprintf('Running BPDN (p=2) reconstruction\n');
[x_bpdn,D_bpdn] = bpdq_1d(yq,A,epsilon2,2,bpdq_opts{:});
Running BPDN (p=2) reconstruction
Examine results
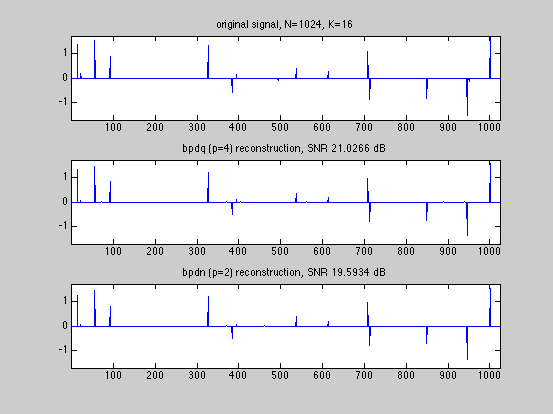
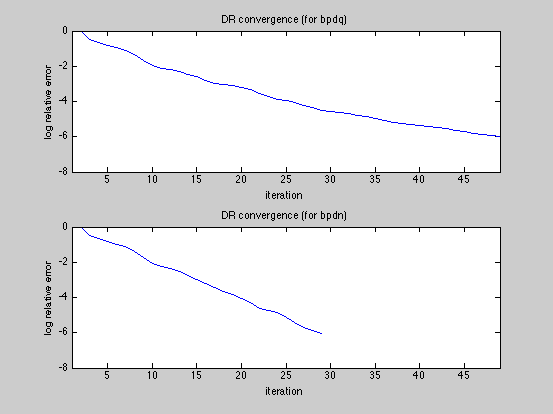
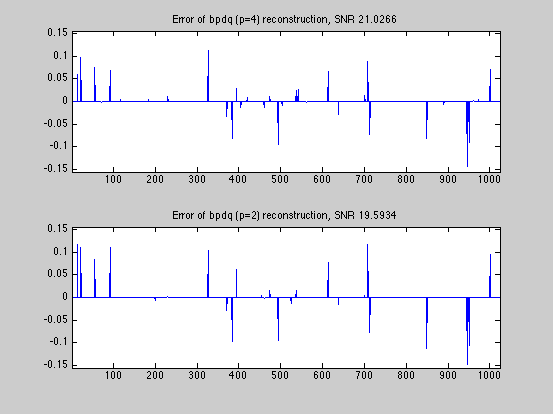
SNR_bpdq = bpdq_compute_snr(x,x_bpdq); SNR_bpdn = bpdq_compute_snr(x,x_bpdn); fprintf('Displaying results and convergence curves\n'); % plot results and convergence curves dmax=1.05*max(abs([x(:);x_bpdq(:);x_bpdn(:)])); fig1ylim=[-dmax,dmax]; fig1xlim=[1,numel(x)]; figure(1) subplot(3,1,1); tstr=sprintf('original signal, N=%g, K=%g',N,K); plot(x); xlim(fig1xlim); ylim(fig1ylim); title(tstr); subplot(3,1,2); tstr=sprintf('bpdq (p=%g) reconstruction, SNR %g dB',p,SNR_bpdq); plot(x_bpdq); xlim(fig1xlim); ylim(fig1ylim); title(tstr); subplot(3,1,3); tstr=sprintf('bpdn (p=2) reconstruction, SNR %g dB',SNR_bpdn); plot(x_bpdn); xlim(fig1xlim); ylim(fig1ylim); title(tstr); figure(2) subplot(2,1,1) fig2xlim=[1,max(numel(D_bpdq.dr_relerr_save),... numel(D_bpdn.dr_relerr_save))]; plot(log10(D_bpdq.dr_relerr_save)); xlim(fig2xlim); title('DR convergence (for bpdq)'); xlabel('iteration'); ylabel('log relative error'); subplot(2,1,2) plot(log10(D_bpdn.dr_relerr_save)); xlim(fig2xlim); title('DR convergence (for bpdn)'); xlabel('iteration') ylabel('log relative error'); figure(3) diff_bpdq=x-x_bpdq; diff_bpdn=x-x_bpdn; dmax=1.05*max([abs(diff_bpdq);abs(diff_bpdn)]); fig3ylim=[-dmax,dmax]; fig3xlim=[1,numel(x)]; subplot(2,1,1); tstr=sprintf('Error of bpdq (p=%g) reconstruction, SNR %g',p,SNR_bpdq); plot(diff_bpdq); xlim(fig3xlim); ylim(fig3ylim); title(tstr); subplot(2,1,2); tstr=sprintf('Error of bpdq (p=2) reconstruction, SNR %g',SNR_bpdn); plot(diff_bpdn); xlim(fig3xlim); ylim(fig3ylim); title(tstr); % The BPDQ Toolbox is free software: you can redistribute it and/or modify % it under the terms of the GNU General Public License as published by % the Free Software Foundation, either version 3 of the License, or % (at your option) any later version. % % The BPDQ Toolbox is distributed in the hope that it will be useful, % but WITHOUT ANY WARRANTY; without even the implied warranty of % MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the % GNU General Public License for more details. % % You should have received a copy of the GNU General Public License % along with The BPDQ Toolbox. If not, see <http://www.gnu.org/licenses/>.
Displaying results and convergence curves