- français
- English
Background coliform bacteria
- Background Coliform Bacteria
Background Coliform Bacteria
http://vulgariz.com/wp-content/uploads/2011/05/escherichia-coli-entero-hemorragique.jpg
Coliform bacteria are Gram-negative bacteria, which can ferment lactose. They are commonly found in nature and in feces of warm-blooded animals. This class of bacteria is used to determine fecal contamination of water, not because they are dangerous (although some are), but because they indicate the presence of other pathogenic organisms of the same origin (like viruses, protozoa and many multicellular parasites). They are also easy to culture.
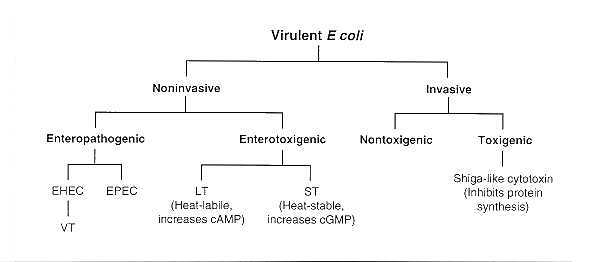
Escherichia Coli (E.Coli) is the most studied member of this familiy and is found in the intestinal tract of many animals including humans. It is mostly harmless, but some strains express virulence factors that are dangerous.
These pathogenic strains are transmitted via oral absorption of fecal-contaminated water for example (they can be found in food too). One of these strains is the Shiga-toxin E.Coli (STEC), which is found in in ruminant guts and is mainly known in the cas of food contamination. This is one example of illness-causing E.Coli strain.
http://web.archive.org/web/20071102062813/http://www.gsbs.utmb.edu/microbook/ch025.htm
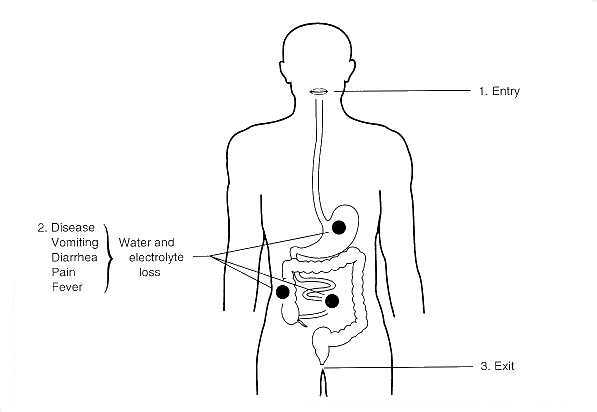
Coliform bacteria intoxication often leads to bloody diarrhea and vomitting (gastroenteritis, dysentry), but some pathogenic strains can cause complications such as hemolytic uremic syndrome (HUS). HUS consists of clot-formation thus blocking arteries and destroying red blood cells, leading to reduced blood flow and ischemia (lack of oxygen and glucose due to a reduced blood supply to tissues).
http://web.archive.org/web/20071102062813/http://www.gsbs.utmb.edu/microbook/ch025.htm
As we just said, identification of coliform bacteria gives indication on fecal contamination of water. For this measure, the approved method is the HPC (heterotrophic plate count), which involves several test conditions depending on what microorganism we want to detect (heterotrophs include bacteria, yeasts and moulds, and are characterized by their need for organic carbon to grow) in addition with other tests. The standard conditions are: temperature (20-40°C), incubation time (few hours to days or even weeks) and nutrient concentration (low/high).
“In water considered safe, this value lies between 0 and 100 cfu/ml.”[1] Studies found that this method may underestimate the quantity of bacteria, thus it is important to continue ameliorating the procedures, as well as developing new methods for water safety control.
The presence of microorganisms is in general determined by high HPC values, which are found in stagnant regions. “In piped water supplies, for example, the HPC is required to be less than 300 colony-forming units (cfu)/ml. The presence of E. coli or enterococci is regarded as evidence of fecal contamination; accordingly, a 100 ml water sample has to be shown to be completely free of E. coli and enterococci.”[2]
To have an overview of what was possible to do, we compiled a table of the latest methods used for E.Coli detection. The criteria we took in consideration were : detection range, time scale, cost and waste. Another important parameter was the additional « hardware » needed to accomplish the different experiments.
Among the methods available, we disthinguised different class :
- PCR-based methods (time consuming)
- Colorimetry (lot of chemicals)
- Bioluminescence
- Florescence – spectrometry
For the PCR methods, we can see that the limiting aspect is the incubation time (several hours). The Colorimetry method requires a lot of chemical reagent that we would have to process, so this raises concern about the waste handling.We have then Bioluminescence and Fluorescence, that both seem to match our criteria. Meaning that they have a quite good detection range and a small time requirement.
The technique that retained our attentionis the Fluorescence, because it matches our criteria, but also because it allows to detect Arsenic as well. It is based on the intrinsic fluorescence of bacteria that originates from their chemical components. Aromatic amino acids (tryptophan, tyrosine, phenylalanine), nucleic acids and co-enzymes are native fluorophores that excite with UV light.
More specifically, if we excite E.Coli at 280-290nm, they show fluorescence in the 300-400nm range, which corresponds to tryptophan residues. E.Coli use Trp to produce indole, which work as extracellular signal.
Go back to top of the paragraph
[1], [2] “New methods for assessing the safety of drinking water, research reports”, T. Egli, Eawag News 65e/December 2008
