|
Navigation
|
SPM PipelineThe SPM pipeline segments the brains into GM/WM/CSF/Other and creates a average template of all tissues. For the purpose of Alzheimer classifier the GM (tissue type 1) is the only tissue of interest. The file "training_SPM_DARTEL_create_template.mat" loads a batch with all the necessary settings to process the segmentation and warping into the average template space. The only thing to do is to add the cases of interest to the “Named File Selector” module. This can be done either by browsing/selecting the files or by use of the “Ed” button by pasting a list of files. Before starting the actual pipeline, you should check the consistency of the data, especially the dimensions of the volume that must be exactly the same for all scans. The simplest way to check the homogenity is by using 'VBM8->Tools->Display one slice for all images'. It is also a good idea to test the pipeline with two or three scans to test if the process runs without errors. During this process a cup of coffee may be approriate ;-) Use a prepared batch as template. For the default settings, only two things have to be configured.
Prepare MATLABBefore starting, please make sure you followed all the steps in the installation section. Start MATLAB and run >> spm
click the "PET & VBM" button
click the "Batch" button
To load the VBM module:
To load spm_pipeline module:
To load a template batch:
Prepare the BatchClick on the module "Change Directory" and select the working directory by double clicking on the item "Directory". Change to the desired directory and click on "Done".
To add the list of files to be processed, there are two different ways: a)
b)
Make sure that in the module NewSegment, the path for the tissue probability maps is correct:
If some path are incorrects, correct them by:
-> in the batch editor, the tissue probability maps should appear in the following format: filepath, tissue type. Ex: D:\Program Files\MATLAB\R2009b\toolbox\spm8\toolbox\Seg\TMP.nii,2 (for WM Note: don't change the number of gaussian from its default value. Run the BatchRun the batch by clicking on the green triangle. The result of the process will be file called "dp_mwc1.mat". The dod product variable name is "Phi". (> load dp_mwc1.mat) |

 (image: spm_start)
(image: spm_start) (image: pet_vbm)
(image: pet_vbm) (image: batch)
(image: batch) (image: batch_vbm)
(image: batch_vbm) (image: batch_vbm_classifier_pipeline)
(image: batch_vbm_classifier_pipeline)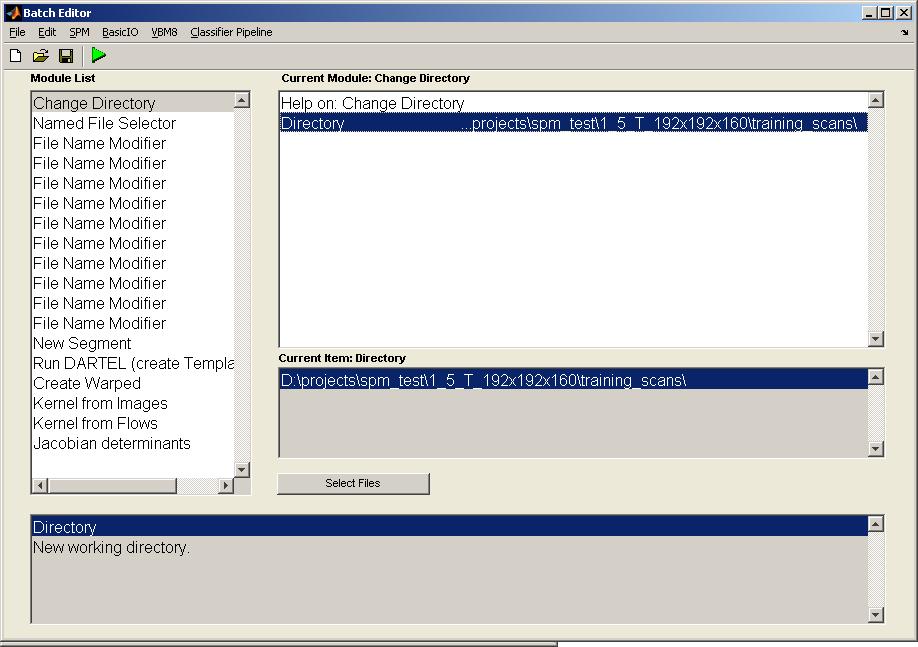 (image: loaded_batch)
(image: loaded_batch)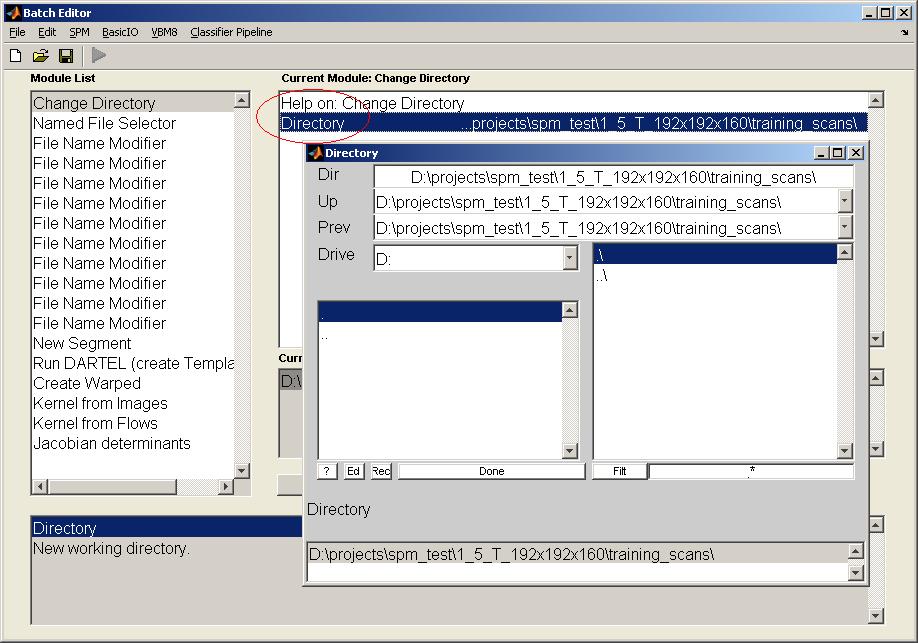 (image: batch_change_directory)
(image: batch_change_directory)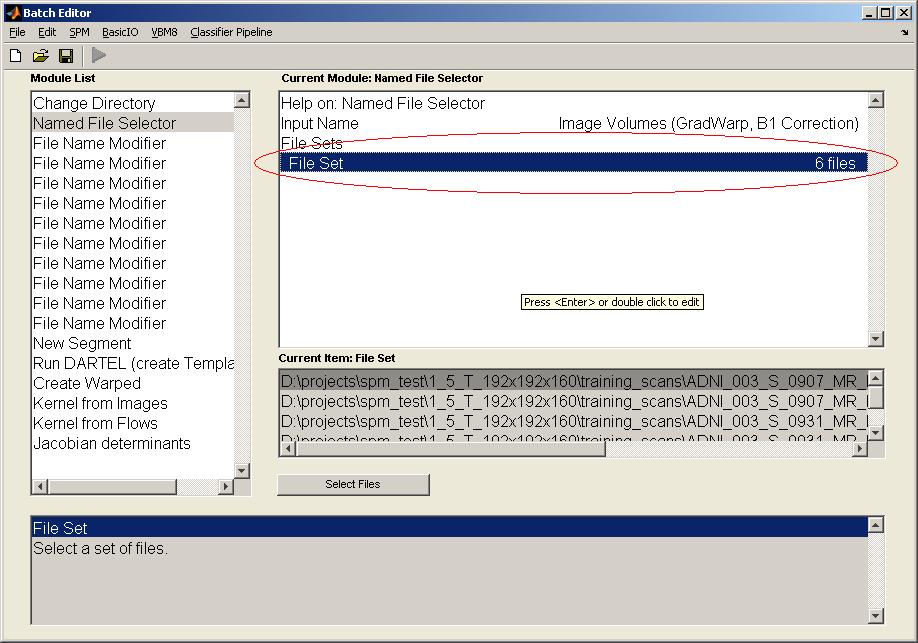 (image: batch_file_set)
(image: batch_file_set)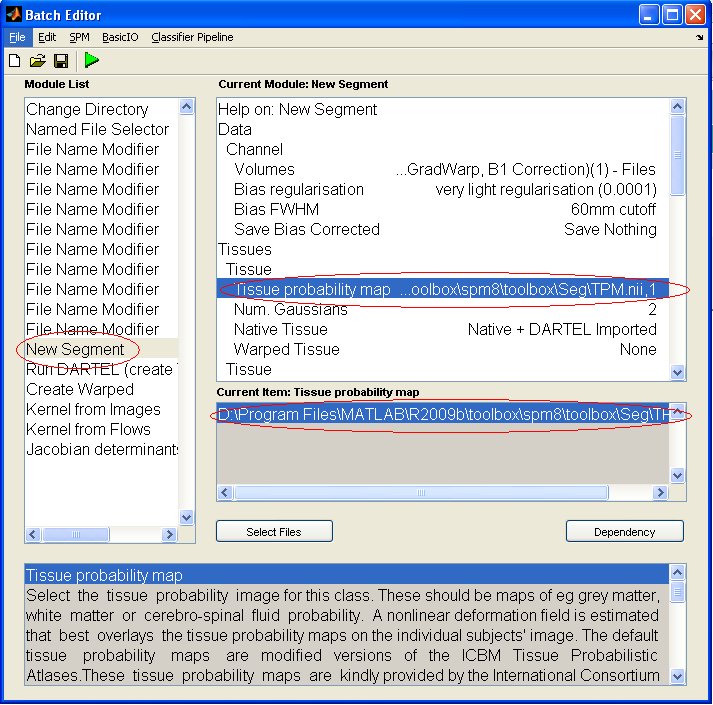 (image: batch_newseg.jpg)
(image: batch_newseg.jpg)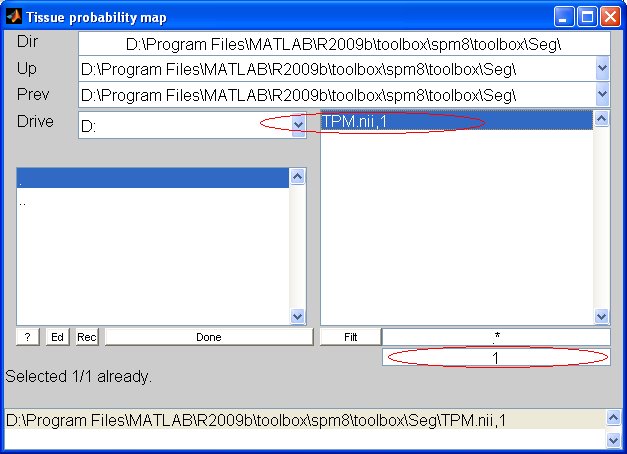 (image: tissue_prob_map.jpg)
(image: tissue_prob_map.jpg) )
)