|
Navigation
|
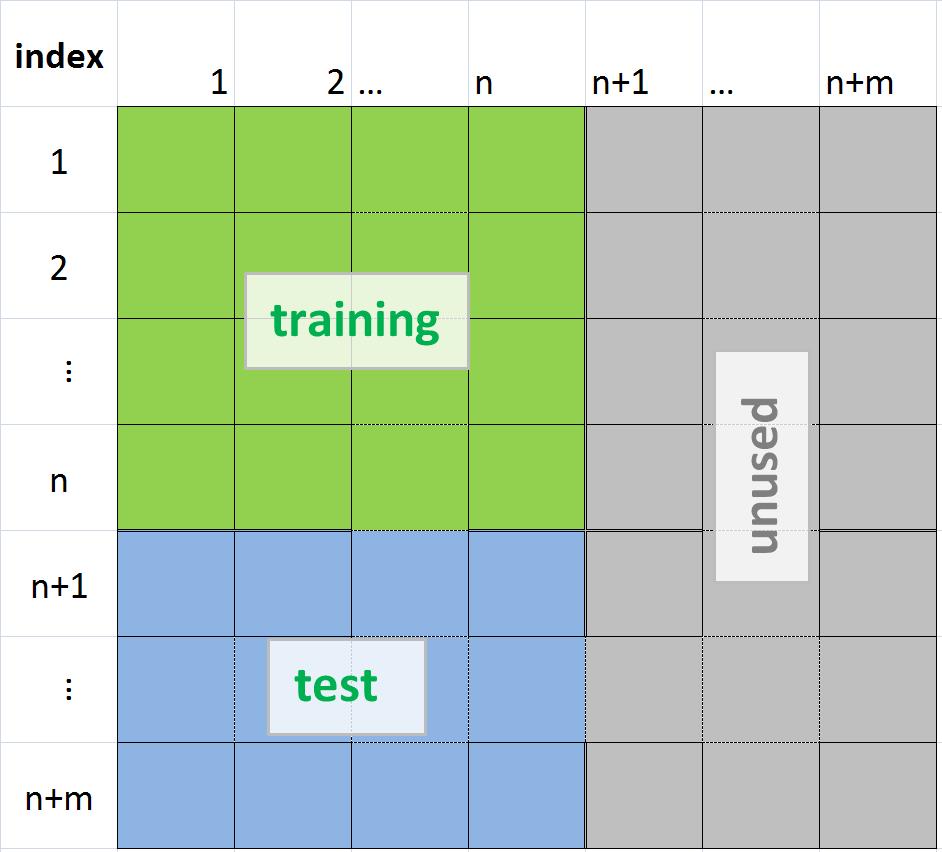
SVMThe SVM has to be trained to find the "best" parameters. This is done by a training set, where the group of each element is known. The estimated parameters are then used to predict the group of an incoming element. To test the training quality one can use a set of elements where the group is known, but which were not used to find the SVM parameters. Before using SVM make sure you have installed Python, gnuplot and libsvm. See installation for a description how to install the software. See this paper for an introduction to libsvm. You can download the software from here. Important: The results depend massively on the parameters. In order do be able to reproduce the result the parameters have to be stored together with the results. The script easy.py does not produce the same result for two different versions of libsvm. It should therefore not be used as reference. The input to the SVM is a dot product matrix. It consists of the n training samples and m testing samples. The green part is used for training (m=0 if no testing data). The blue part is used for testing the obtained parameters. See the sections "Training the SVM" and "Testing the SVM" how to extract the values from the matrix (MATLAB variable Phi) into a format that libsvm can read. Training the SVM (automated) The input file format of SVM has following format (assuming m=0)
Example:
Depending on the configuration of the SPM, the output file is called something like dp_mwrc1.mat. Load this file into matlab >> load(/path/to/dp_mwc1.mat) To get a file formatted in the way like described above try something like this
To apply a the SVM on the dot product matrix, change to the subfolder tools/ of the libsvm-2.X directory and run. python easy.py /path/to/dp_matrix/phi_svm.data Information: To run the easy.py script outside the tools subfolder you need to change the script itself. Edit: Alternatively you can use this script to generate a training and testing data file. The Python scripts guesses an ideal "C"-value (see the paper mentioned above) and does a cross validation with the given data set. Testing the SVM (automated)To test the parameters you need to generate a new dot product matrix. Assuming the testing dp-matrix was n-by-n and you add m testing cases, the resulting matrix will be (n+m)-by-(n+m). The testing file, let's call it 'phi_svm_test.data' will have the columns 1-n and the rows (n+1)..(n+m). The format is the same as for the file 'phi_svm.data'. When changing the indexes, one can use the script above to generate the file. The simple way to train and test a given data set is to execute. Change: for i=1:size(Phi,1); -> for i=n+1:size(Phi,1); Change: for j=1:size(Phi,1); -> for j=1:n; run on command line: python easy.py phi_svm.data phi_svm_test.data For more details, see the libsvm-guide. The accuracy of the test samples is in general smaller than the accuracy of the training set. Training and Testing with Manual ChangesThe approach described above uses a script and default values to train the machine. However, the training can be influenced by a various parameters such as the kernel, the scaling and the parameters C and gamma. See the libsvm-guide for a description of the possibilities. Output FilesThe easy.py script creates some files (assuming the imputs are training.data and test.data)
|