- français
- English
GIF model parameter extraction
In this step, the experimental data will be used to fit a Generalized Integrate-and-Fire (GIF) model equipped with a spike-triggered current and a spike-triggered movement of the firing threshold (see Figure).
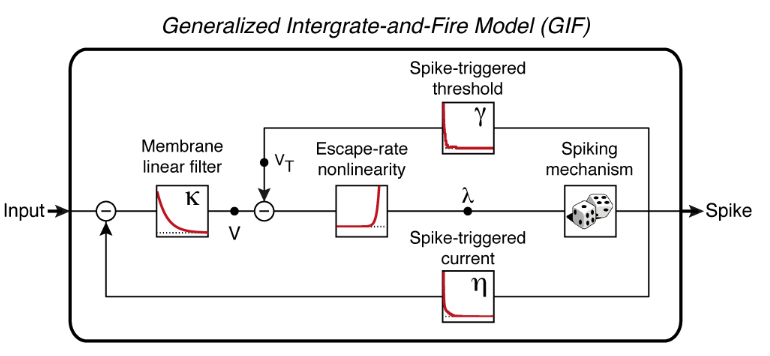
Figure: schematic representation of the Generalize Integrate-and-Fire (GIF) model. The input current is first passed through the membrane filter and then transformed into a spiking probability (lambda) by the escape-rate nonlinearity. Spikes are produced stochastically and triggers both an adaptation current and a threshold movement. V: membrane potential. VT firing threshold. Further details on the GIF model can be found in the original manuscript.
1. Import the class GIF and create a new object myGIF
To create the object, specify the time step dt (in ms) that will be used for numerical integration of the GIF model (this parameter does not necessarily have to be the experimental sampling frequency).
from GIF import *
myGIF = GIF(0.1)
2. Specify the absolute refractory period
This parameter is not extracted from the data, but has to be specified. For pyramidal neurons 4 ms is a good choice. In general, the absolute refractory period should be much smaller than the shortest interspike-interval in the data. The parameter has to be specified in ms.
myGIF.Tref = 4.0
3. Define the rectangular basis functions used for the filters eta and gamma
According to Equations 16 and 19 (Pozzorini et al. 2015), the spike-triggered current and the spike-triggered threshold movement are expanded using rectangular basis functions. To use log-spaced basis functions use the following code:
from Filter_Rect_LogSpaced import *
myGIF.eta = Filter_Rect_LogSpaced()
myGIF.eta.setMetaParameters(length=5000.0, binsize_lb=2.0, binsize_ub=1000.0, slope=4.5)
myGIF.gamma = Filter_Rect_LogSpaced()
myGIF.gamma.setMetaParameters(length=5000.0, binsize_lb=5.0, binsize_ub=1000.0, slope=5.0)
The parameter length (in ms) defines the total duration of the filter. The parameters binsize_lb and binsize_ub define the size (in ms) of the smallest and of the largest rectangular basis function. Finally, the parameter slope defines the scaling exponent at which the width of basis functions increases.
For linearly spaced rectangular basis functions use the class Filter_Rect_LinSpaced. For arbitrarily spaced rectangular basis functions use the class Filter_Rect_ArbitrarilySpaced. To define a new set of basis functions, write a new class which inherits from Filter.
4. Fit the experimental data in myExp using the object myGIF
To fit the GIF model myGIF to the experimental data in myExp use the following command:
myGIF.fit(myExp, DT_beforeSpike=5.0)
Note that only the data defined as training set will be used for the fit. Since the GIF model does not capture the subthreshold dynamics of the membrane potential during spike initiation, the data preceding an action potential are discarded. Use the parameter DT_beforeSpike to specify (in ms) the region that has to be discarded before each spike.
To perform the fit using only a specific part of the training set, use the following command before calling myGIF.fit():
myExp.trainingset_traces[0].setROI([[0,10000.0], [20000.0, 60000.0]])
This function defines a Region Of Interest (ROI) between 0 and 10000 ms and between 20000 and 60000 ms. Selected ROIs can be visualized by calling the function myExp.plotTrainingSet().
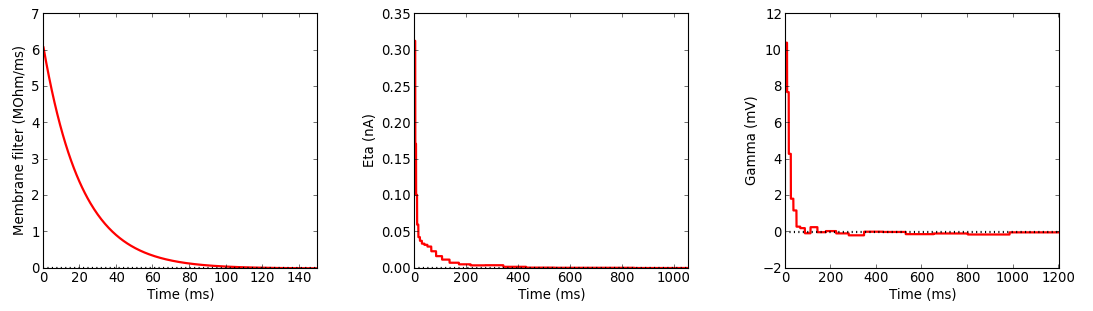
5. Visualize GIF model parameters and save the results
To print (on the terminal) the GIF model parameters and to plot the GIF model filters, use the following functions:
myGIF.printParameters()
myGIF.plotParameters()
To save and reload the GIF model use the following functions:
myGIF.save('./myGIF.pck')
myGIF_reloaded = GIF.load('./myGIF.pck')
To simulate the model response to a current I with initial condition V(t=0)=V0, use the following function:
(time, V, I_a, V_t, S) = myGIF.simulate(self, I, V0)
The function returns 5 arrays:
- time: support that can be used to plot the other quantities (in ms)
- V: voltage trace (in mV)
- I_a: adaptation current (in nA)
- V_t: voltage threshold (in mV)
- S: spike times (in ms)
In the last step, the myGIF model will be used to predict the spiking activity in the test dataset of myExp.
